The ssWavelets Package: Wavelet Functionality for Package sampSurf
Overview of the ssWavelets package
The
In general, the wavelet filter decomposes the sampSurf simulation results by
scale (distance), with each scale corresponding to a different level of the
decomposition. This results in a set of anisotropic wavelet coefficients for
each scale or level, one each for horizontal, vertical and diagonal components; the
sum of these yields an isotropic decomposition for that
scale. Residual smooth coefficients for the coarsest level are also available
from applying the scaling filter, and its mean corresponds to the sample mean
of the sampling surface from sampSurf. Interesting as these
Details of package use are found in the package vignette:
ssWavelets installation
Before installing
and can include the
In addition, the
ssWavelets package vignette
As noted above, there is one extra package vignette (aside from the default help vignette) distributed with the package itself. Both can also be downloaded here from the R-Forge versions if desired…
The following gives another simple application of wavelets to sampling surfaces…- Gove, J. H., 2017. Some Refinements on the Comparison of Areal Sampling Methods via Simulation. Forests 8, 393:1-24; doi:10.3390/f8100393.
ssWavelet examples
Here we present one very simple example illustrating how to use a few of the main
functions in the
Critical Height Sampling
The following code will generate a volume sampling surface using critical height sampling
(CHS) in the form of a
R> require(sampSurf) R> tract.m = Tract(c(x = 64, y = 64), cellSize=1) #square tract, dyadic grid ~0.5ha R> buffTr.m = bufferedTract(bufferWidth = 10, tract.m) R> ag3m = angleGauge(3) #metric R> sschs = sampSurf(25, buffTr.m, iZone = 'criticalHeightIZ', angleGauge = ag3m, dbhs = c(12,30), + topDiam = c(0,0), startSeed = 304, referenceHeight = 'dbh')
Number of trees in collection = 25 Heaping tree: 1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,
R> plot(sschs, useImage = FALSE)
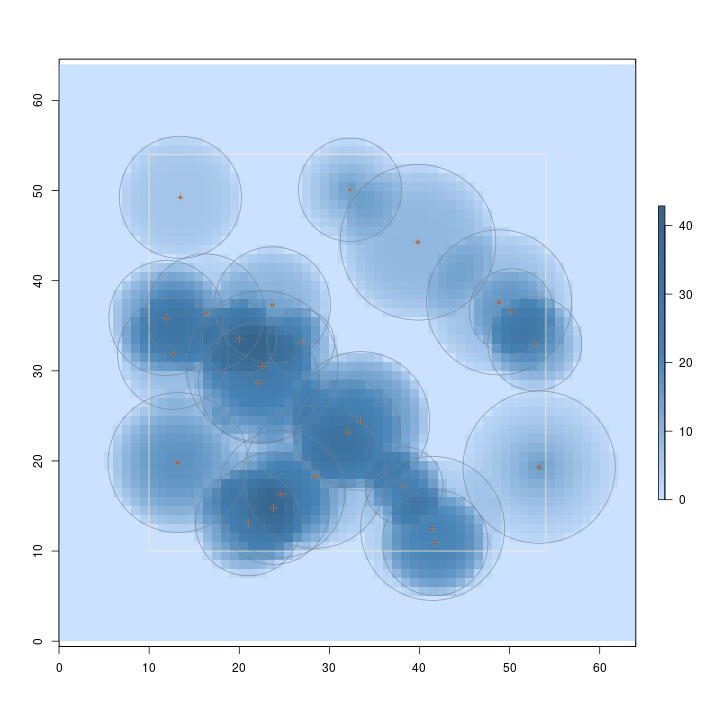
The above figure presents the volume sampling surface for CHS. Note how the surface (volume estimate) varies within each tree's inclusion zone, depending on its dimensions.
Now we can do a MODWT analysis on this surface…
R> J.chs = 5 #highest decomposition level: J_0 R> chs.modwt = ssMODWT(sschs, J = J.chs)
Inclusion zone class: criticalHeightIZ Surface variance = 75.792717 Wavelet variance = 75.774213 (MODWT) Surf/Wave var ratio = 1.0002442 Check: surface var matrix = 75.774213 = E[X^2] - E[X]^2 Surface mean = 6.0179401 Wavelet mean = 6.0179401 (MODWT) Wavelet mean = 6.01794 (MRA)
R> plotLevel2D(chs.modwt, type = 'var', level = 1, decompType = 'modwt')
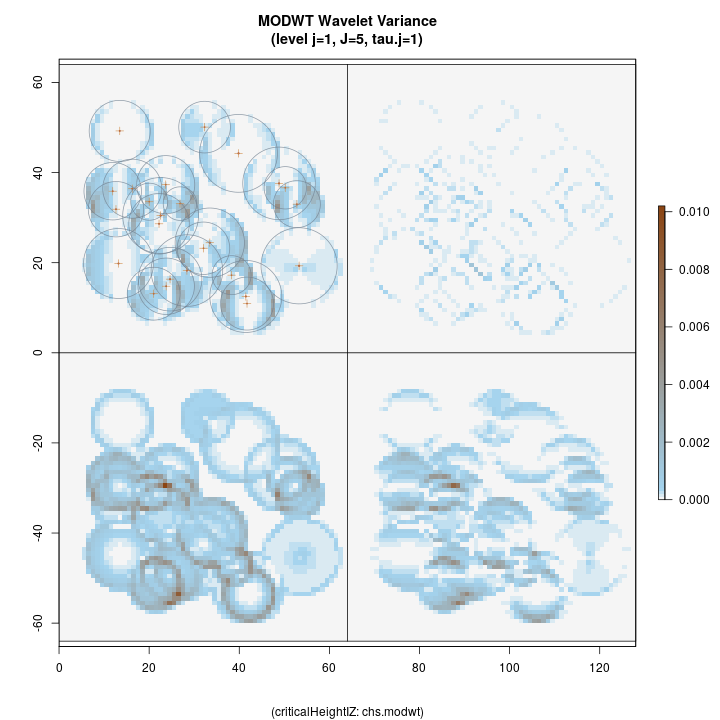
Top left: horizontal Top right: diagonal Bottom right: vertical Botton left: isotropic
The figure above shows the four anisotropic variance decompositions at
level j=1, corresponding to the legend printed below the figure. This
first-level decomposition is good for
The raw wavelet coefficients are shown in the following figure for comparison. Here we can see how positive coefficients are generated as the wavelet encounters the boundary of an inclusion zone coming from a background value (of zero), or more generally as the surface increases in the direction of the wavelet filter. Conversely, negative values are generated as the surface decreases in the same direction as the filter.
R> plotLevel2D(chs.modwt, type = 'raw', level = 1, decompType = 'modwt')
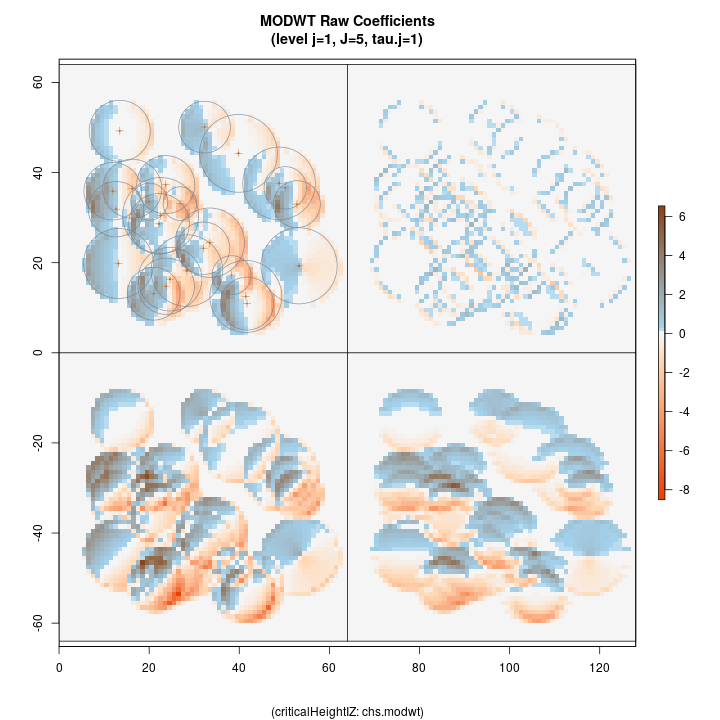
Top left: horizontal Top right: diagonal Bottom right: vertical Botton left: isotropic
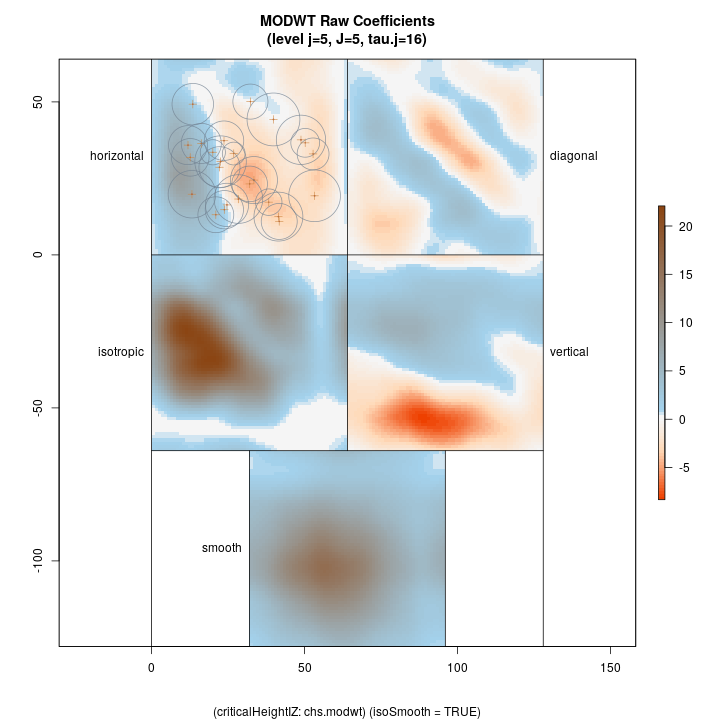
Lastly, we show the coarsest level decomposition for both the variance and raw coefficients, respectively. Note that the final image is of the smooth; these images show larger scale relations…
R> plotLevel2D(chs.modwt, type = 'var', level = J.chs, decompType = 'modwt')
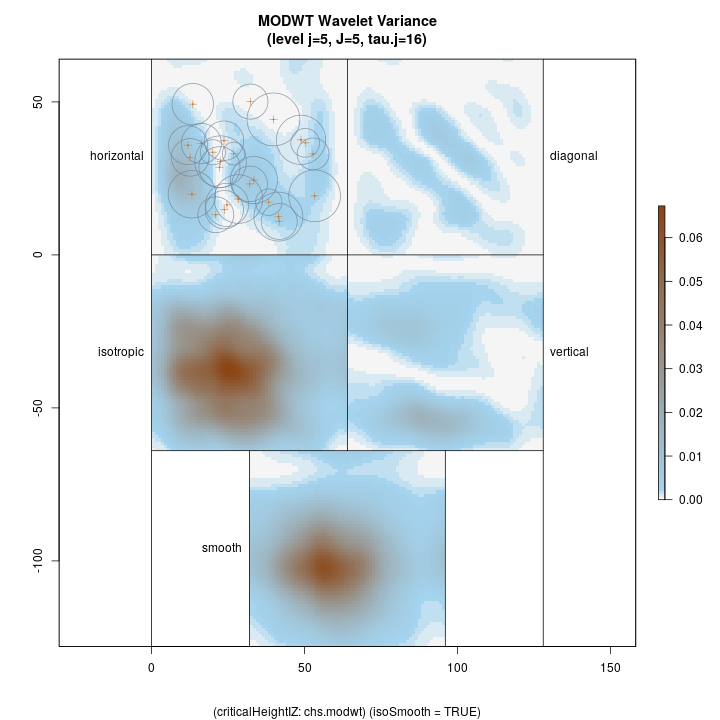
Top left: horizontal Top right: diagonal Bottom right: vertical Botton left: isotropic Bottom middle: final smooth
R> plotLevel2D(chs.modwt, type = 'raw', level = J.chs, decompType = 'modwt', runQuiet = TRUE)
Summary
There are many more aspects to this package as outlined in the User's Guide. These include plotting functions that allow the comparision of different sampling methods by scale and covariance analysis. It also describes the edge or boundary correction methods available in the wavelet analyis phase.
